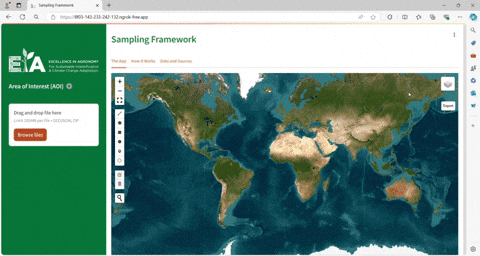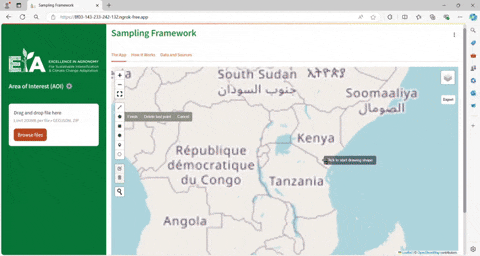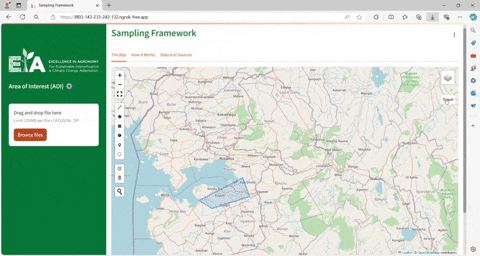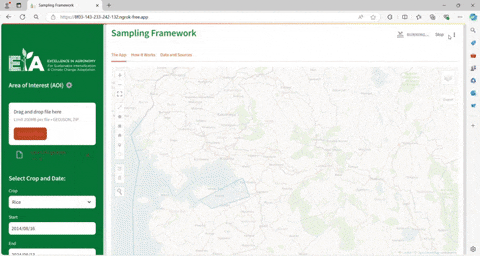
Seasonal conditions affect how environments might be clustered within the AOI, thus affecting the results
of the trials and how the crop performs. The tool requires that the user indicates the seasonality (start month and end month)
when the trials will be conducted.
where the user will be able to create a more tailored set of clusters.
-
5. Relevant environmental variables
+
5. Relevant environmental variables
At this point, the user may also decide which are the most relevant environmental variables that may
influence the trials or that are of specific focus in the validation. For example, an MVP might be focusing on drought-resistant
varieties in rainfed wheat, and therefore rainfall might be the most important environmental variable to define the
diff --git a/SFapp.py b/SFapp.py
index 2636e98..a317f93 100644
--- a/SFapp.py
+++ b/SFapp.py
@@ -32,7 +32,8 @@
'Temperature Minimum','Temperature Mean','Soil Zinc', 'Elevation','Slope', 'Soil Organic Carbon','Soil pH','Soil CEC','Soil Nitrogen','Soil Clay','Soil Sand']
cropmask = ee.Image('COPERNICUS/Landcover/100m/Proba-V-C3/Global/2019').select('discrete_classification').eq(40)
-Map = geemap.Map(center=[0, 0], zoom=2, Draw_export=True)
+Map = geemap.Map(center=[0, 0], zoom=2, Draw_export=True)
+Map.add_basemap("SATELLITE")
# Load CSS file content
with open('style.css', 'r') as css_file:
@@ -51,8 +52,8 @@
with st.sidebar:
st.image(logo)
# st.experimental_rerun()
- st.write("## Upload AOI :gear:")
- data = st.file_uploader("Upload AOI Shapefile", type=["geojson","zip"])
+ st.write("## Area of Interest (AOI) :gear:")
+ data = st.file_uploader(" " ,type=["geojson","zip"])
if data is not None:
st.write("## Select Crop and Date: ")
@@ -101,6 +102,7 @@
# Create map
Map = geemap.Map(center=centroid_coords, zoom=8)
+ Map.add_basemap("SATELLITE")
Map.add_gdf(poly, 'AOI')
#Map.addLayer(bb, {}, 'Bounding Box')
selected_Sdate1 = (datetime.date.today() - datetime.timedelta(days=10*365))
@@ -158,7 +160,7 @@
bb_clip = mapping(poly.geometry.unary_union)
res = res.clip(bb_clip)
- # numClusters = numClusters.getInfo()
+ numClusters = numClusters.getInfo()
# def gen_colors(num_colors):
# random.seed(0) #ensure reproducibility
@@ -296,6 +298,7 @@ def download_file(file_path):
Map = geemap.Map(center=[0, 0], zoom=2, Draw_export=True)
Map = geemap.Map(center=centroid_coords, zoom=8)
+ Map.add_basemap("SATELLITE")
Map.add_gdf(poly, 'AOI')
Map.addLayer (res.randomVisualizer(), {}, 'Kmeans')
@@ -374,7 +377,7 @@ def download_file(file_path):
except Exception as e:
st.error(e)
# st.error("An error occurred: There is an issue with your file download. Try again")
-
+
Map.to_streamlit(height=600)
@@ -407,9 +410,9 @@ def download_file(file_path):
'Rainfall Total': {'property': 'prcSum', 'unit': 'mm'},
'Rainfall Days': {'property': 'prcNrd', 'unit': 'days'},
'Rainfall Average': {'property': 'Di', 'unit': 'mm/day'},
- 'Temperature Maximum': {'property': 'tmaxMax', 'unit': '°C'},
- 'Temperature Minimum': {'property': 'tminMin', 'unit': '°C'},
- 'Temperature Mean': {'property': 'tmeanMean', 'unit': '°C'},
+ 'Temperature Maximum': {'property': 'tmaxMax', 'unit': 'K'},
+ 'Temperature Minimum': {'property': 'tminMin', 'unit': 'K'},
+ 'Temperature Mean': {'property': 'tmeanMean', 'unit': 'K'},
'Soil Zinc': {'property': 'zinc', 'unit':' ppmg'},
'Elevation': {'property': 'srtm', 'unit': 'm'},
'Slope': {'property': 'slp', 'unit': '%'},
diff --git a/__pycache__/eefun.cpython-311.pyc b/__pycache__/eefun.cpython-311.pyc
index dcc148f..dc626f2 100644
Binary files a/__pycache__/eefun.cpython-311.pyc and b/__pycache__/eefun.cpython-311.pyc differ
diff --git a/datasets.html b/datasets.html
index 38b2a13..ac689c8 100755
--- a/datasets.html
+++ b/datasets.html
@@ -705,10 +705,10 @@
Datasets
1979 – Present
-ºC
+K
|
-Minimum temperature (ºC) over the selected period in the target region.
+Minimum temperature (K) over the selected period in the target region.
|
@@ -728,10 +728,10 @@ Datasets
1979 – Present
-ºC
+K
|
-Maximum temperature (ºC) over the selected period in the target region.
+Maximum temperature (K) over the selected period in the target region.
|
@@ -751,10 +751,10 @@ Datasets
1979 – Present
-ºC
+K
|
-Average temperature (ºC) over the selected period in the target region.
+Average temperature (K) over the selected period in the target region.
|
diff --git a/style.css b/style.css
index 163f2c6..8892d47 100644
--- a/style.css
+++ b/style.css
@@ -1,3 +1,87 @@
+/* Theme details*/
+.st-emotion-cache-13k62yr {
+ position: absolute;
+ background: #fff;
+ color: #ccc;
+ inset: 0px;
+ color-scheme: dark;
+ overflow: hidden;
+}
+.st-emotion-cache-h4xjwg {
+ position: fixed;
+ top: 0px;
+ left: 0px;
+ right: 0px;
+ height: 3.75rem;
+ background: rgb(255 255 255 / 0%);
+ outline: none;
+ z-index: 999990;
+ display: block;
+}
+
+.st-emotion-cache-1itdyc2 {
+ position: relative;
+ top: 2px;
+ background-color: rgb(0, 104, 56);
+ z-index: 999991;
+ min-width: 244px;
+ max-width: 550px;
+ transform: none;
+ transition: transform 300ms, min-width 300ms, max-width 300ms;
+}
+
+.st-emotion-cache-xwtqgq {
+ display: flex;
+ -webkit-box-align: center;
+ align-items: center;
+ padding: 1rem;
+ background-color: #fff;
+ border-radius: 0.5rem;
+ color: rgb(49, 51, 63);
+}
+
+
+.st-emotion-cache-7oyrr6 {
+ color: #ccc;
+ font-size: 14px;
+ line-height: 1.25;
+}
+
+.st-emotion-cache-lpgk4i {
+ display: inline-flex;
+ -webkit-box-align: center;
+ align-items: center;
+ -webkit-box-pack: center;
+ justify-content: center;
+ font-weight: 400;
+ padding: 0.25rem 0.75rem;
+ border-radius: 0.5rem;
+ min-height: 2.5rem;
+ margin: 0px;
+ line-height: 1.6;
+ color: #f8f9fa;
+ width: auto;
+ user-select: none;
+ background-color: rgb(195, 82, 31);
+ border: 1px solid rgba(250, 250, 250, 0.2);
+}
+
+.st-ci {
+ background-color: #fff;
+}
+.st-c1 {
+ color: #333;
+}
+
+::-webkit-scrollbar {
+ background: #ddd;
+ border-radius: 100px;
+ height: 6px;
+ width: 10px;
+}
+
+
+
/* Header styles */
.st-emotion-cache-1r4qj8v {
position: absolute;
@@ -49,6 +133,37 @@
align-items: center;
}
+/* adjusted padding */
+.st-emotion-cache-1jicfl2 {
+ width: 100%;
+ padding: 1rem 1rem 10rem;
+ min-width: auto;
+ max-width: initial;
+
+}
+.st-emotion-cache-12fmjuu {
+ position: fixed;
+ top: 0px;
+ left: 0px;
+ right: 0px;
+ height: 3.75rem;
+ background: rgb(255, 255, 255, 0);
+ outline: none;
+ z-index: 999990;
+ display: block;
+}
+.st-emotion-cache-ato1ye {
+ position: fixed;
+ top: 0px;
+ left: 0px;
+ right: 0px;
+ height: 3.75rem;
+ background: rgb(255, 255, 255, 0);
+ outline: none;
+ z-index: 999990;
+ display: block;
+}
+
/* button */
.st-emotion-cache-13ejsyy:hover {
border-color: rgb(255, 75, 75);
@@ -197,4 +312,6 @@ st.sidebar {
.no-print {
display: none;
}
+}y: none;
+ }
}
\ No newline at end of file