Easily build graphs from Eppendorf - DASGIP MultiparallelBioreactorSystem generated .csv files.
A graphical web interface for generating graphs for DASGIP MBS output. Configuration of limits and colors are saved persistently. Uploaded data and generated graphs are stored for a period of time. The app works as a locally hosted website using Flask. Graphs are generated using matplotlib with a non-GUI backend.
This project was requested to help improve the efficiency of scientists working with the DASGIP MBS and is open to other research groups and companies.
- Python 3.8.x
- Flask 2.3.2
- Flask-Dropzone 1.6.0
- matplotlib 3.7.1
- numpy 1.24.4
- pandas 2.0.3
pip install -r requirements.txt
There are some options for using the program:
DASGIPGraphBuilderis a simple CLI.
- From the main folder, run:
python -m src.DASGIPGraphBuilder <FILENAME> [-s vessel_number] [-d variable, [...]]
appis a Flask web server version.
- From the main folder, run:
python -m src.app
- If no file is available, the main page will redirect to upload automatically.
-
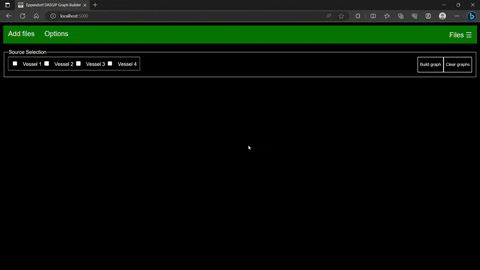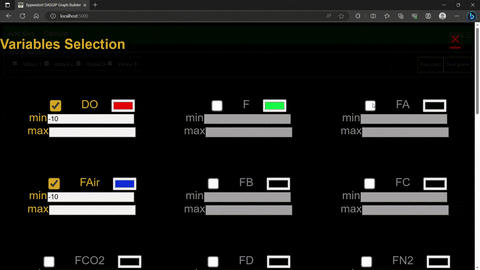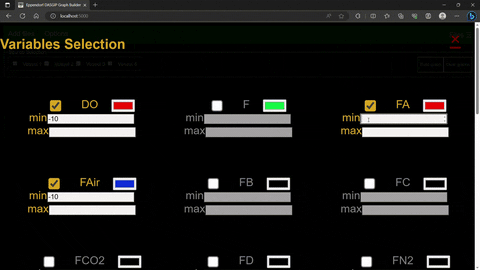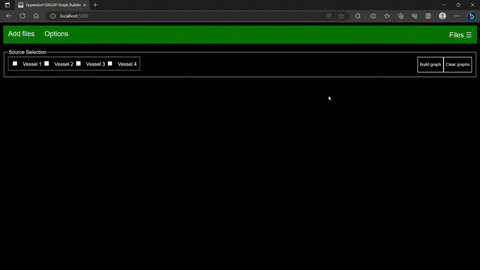
From the main page files, sources within files and options can be selected.
-
The options menu allows for the selection of variables, their color scheme and plot limits.
After uploading files and setting prefered options, graphs are easily generated at the click of a button.
run.shis a bash file that creates a virtual environment on first use and runs the web server in it.
From the main folder, run:
./run.sh
0. Improve graphics for portability.
1. Include admin interface for configuring app behaviour.
2. Add more graph configurations options.
3. Load from compressed file.
* conf.py contains constants and sets logging.
* protocols.py contains interfaces expected by modules.
1. DASGIP_loader:
Implements all tasks related to the internal organization of the CSV.
2. graph_maker:
Implements all graph related operations.
3. handler:
Interface for building graph from file and configuration
settings using a loader module and a grapher module.
4. os_ops:
Implements all OS and time related tasks.
Implements presistent memory as a json file to:
- Store only relevant data content from the file;
- Exclude data entries after configurable time;
- Exclude generated images after configurable time;
- Store graph configuration settings.
1. DASGIPGraphBuilder:
A simple CLI using argparse.
2. app:
A local web server application using Flask.
1 - The program must run locally on a Mac and not depend on remote assets or servers.
2 - The program must create graphs for selected variables in subplots for each file/vessel.
3 - Graphs must be easily configured for color and y-axis limits.
4 - Graphs are subject to tinkering and subjective evaluation.