-
-
Notifications
You must be signed in to change notification settings - Fork 426
New issue
Have a question about this project? # for a free GitHub account to open an issue and contact its maintainers and the community.
By clicking “#”, you agree to our terms of service and privacy statement. We’ll occasionally send you account related emails.
Already on GitHub? # to your account
WAIC/LOO for models with multiple observed variables #987
Comments
* update to numpyro 0.2.1 * update cookbook * fix lint erros * make the test more throughout * no need for divergin field * cleanup notebook file
|
I'd very much like to help get this fixed, if possible, since I have models with multiple observation variables. I'm working in PyMC3, not pyro. Am I correct in thinking that for multiple observations, we need to find the log likelihood of observations that are tuples of individual observed RVs? I don't believe that this operation would be supported natively by PyMC3, but it should be able to add it. Maybe someone with a better understanding of the underlying mathematics could lay out a plan for this? |
I did not get from your description whether you want to leave out If latter, then note that if each |
@rpgoldman Indeed: @avehtari Thank you for the links. Are you saying that WAIC and PSIS often fail in the case where each datapoint |
It's bit difficult that you are using different words than what is common in statistics. For me |
|
Here is an example of a simple model. For an input To compute WAIC/PSIS-LOO for this model, we need all the My question is: shouldn't we simply compute the products of the likelihoods, or equivalently sum all the log_likelihoods? Indeed, If the answer is yes, isn't it something which can be expanded to more complicated models with y1, ..., yN, as long as y1, ...yN are independent conditionally on the If the answer is still yes, can it be expanded to models where y1, ..., yN are not independent conditionally on the samples @avehtari From what I saw in your lectures, I think it means we do leave-one-observation-out but the observation is a vector I hope it makes things clearer. |
|
It's more clear now.
WAIC/PSIS-LOO can be computed with either
Correct, if you want to focus on
Yes. There can be computational problem if you have
Yes. Depending on the focus you may consider leave-future-out as in time series (see, e.g. https://arxiv.org/abs/1902.06281) |
|
I think we should have multiple options for different methods. LFO needs also possibility to resample (there are code to do this in pystan, but we really need to refine InferenceData class to enable this). We should have same default as in loo2. |
@avehtari Why? Would the computational problem appear during the computation of the WAIC/PSIS-LOO based on |
|
Anyway, all this means that in the case of multiple observation sites, then we can compute WAIC or PSIS-LOO by initially summing the log_likelihood of all observation sites, as proposed here. In my opinion, it should be the default, rather than having a WAIC/PSIS-LOO for each observation site. |
|
#998 is a simple sub-problem here: trying to figure out if I can work around this limitation for the case of conditionally independent subsets of observations. |
|
I wrote one example on handling multiple observed data in PyMC discourse. https://discourse.pymc.io/t/calculating-waic-for-models-with-multiple-likelihood-functions/4834/5 It is built on top of a PyMC3 model, but the main focus is on ArviZ usage and how to handle the datasets in |
Somehow I had missed this question, but got email from the last comment. PSIS-LOO and WAIC fail if the posterior changes a lot. If we remove all observations directly related to some parameter then the posterior of that parameter is the same as the prior which is likely to much different to the posterior. |
|
|
|
#1616 will take care of some immediate fixes, the end goal however should be to allow computing loo outside compare and pass the result to compare instead of being forced to pass inferencedata to it |
|
Update after 10 days: Dear all, This conversation is very relevant to the problem I am trying to solve, so I joined in and seek some help. I am trying to convert HDDM model data into HDDM is for behavioral experiments in psychology, where human subjects/participants are asked to response to stimuli appeared on computer screen by pressing (one of two) buttons. We call each presentation of a stimulus (and participants' response) as a For each condition, we usually have multiple trials, but the number of trials may not be the same for different conditions, which means that we may have different number of observed data points for different conditions. Worse, because participants may miss some trials, these trials are usually removed from analysis, so that different participants may also have different observed data point under that same condition. For example, in the data I am working with, the data looks like below: if we look at the number of trials of each condition for each participant, it looks like this: As we can see above, for the condition When convert data and point-wise log likelihood to InferenceData, I set the I've read this very valuable post: https://discourse.pymc.io/t/calculating-waic-for-models-with-multiple-likelihood-functions/4834/5 but have no idea how to translate the idea in that post to our situation. I an new to Thanks in advance. PS: if I trim the data so that each participant and each condition has same number of trials, then it works: #1733. |
|
@hcp4715 I have a similar issue as the one you report, have you by any chance made progress on this? It would be super helpful! |
|
Hi, @FrancescPoli , I've updated my original post. Briefly, when converting dataframe to Xarray, Xarray will auto fill the rows with Meaning, the See here for my update: https://groups.google.com/g/hddm-users/c/cO9EBdRAvzs/m/DBg_n4s7BAAJ. |
|
Closing this as the original issue has been fixed. For usage and interpretation questions on loo/waic, please ask on stan or pymc discourse forums. |



(Related issue: #794 - cc @rpgoldman)
First, thank you for this great library!
I was told by @ahartikainen here that multiple observations were not supported in arviz (at least for numpyro and pyro, but I guess for other libraries like PyMC3 as well).
My question is: to go from 1 observation to multiple observations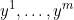 , isn't it enough to sum all the log_likelihoods of observed variables to compute the waic and loo?
, isn't it enough to sum all the log_likelihoods of observed variables to compute the waic and loo?
Indeed, the WAIC and LOO are computed based on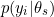 where
where 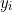 are all the examples (observations) and
are all the examples (observations) and 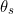 are all the samples from the posterior of all parameters and hidden variables.
are all the samples from the posterior of all parameters and hidden variables.
Below I write examples of numpyro models:
for one observation:
for several observations which are conditionally independent:
for several observations which are not conditionally independent:
The text was updated successfully, but these errors were encountered: