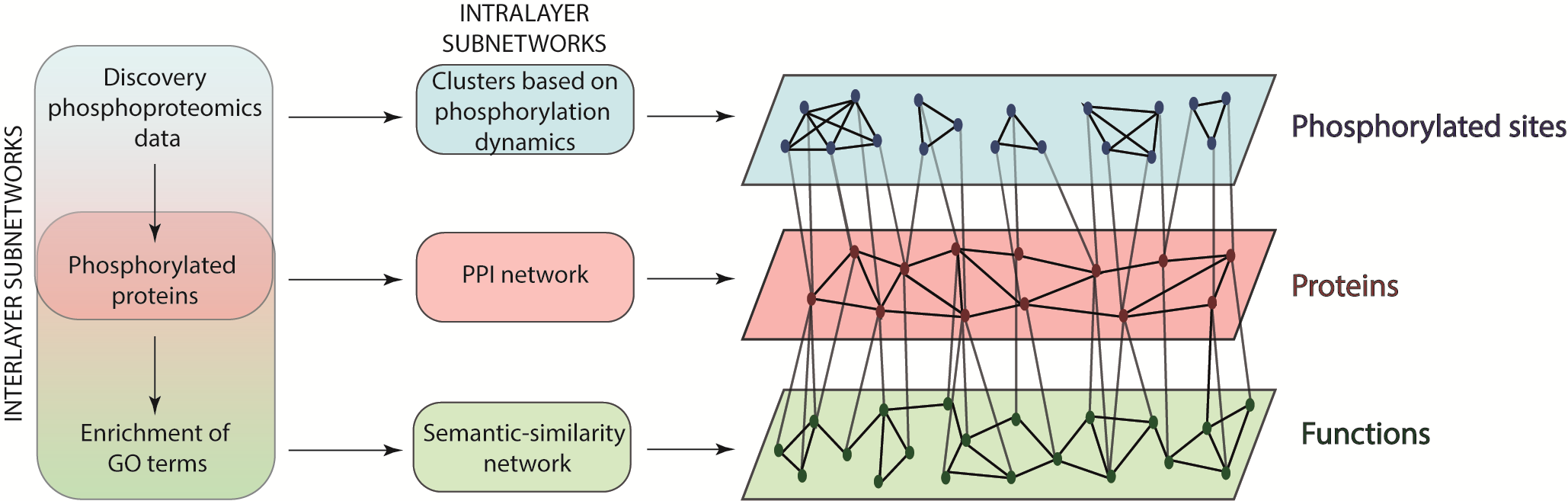
A method to infer the function of individual phosphorylation sites based on phosphoproteomics data, using random walks on heterogeneous networks (RWHN).
Run the following in an R session:
devtools::install_github("JoWatson2011/phosphoRWHN", build_vignettes = T)
The Bioconductor packages org.Hs.eg.db, GOSemSim, GO.db, AnnotationDbi are also required and may need installing separately.
Run the following in an R session:
library(phosphoRWHN)
vignette("phosphoRWHN_MAPKERK")
Manuscript can be viewed here: Watson, Joanne, Jean-Marc Schwartz, and Chiara Francavilla. “Using Multilayer Heterogeneous Networks to Infer Functions of Phosphorylated Sites.” Journal of Proteome Research 20, no. 7 (July 2, 2021): 3532–48. https://doi.org/10.1021/acs.jproteome.1c00150.