-
Notifications
You must be signed in to change notification settings - Fork 6
Database Querying
The relational database component allows efficient monitoring and summarising the output of screening. Particularly when the screening project database is enriched with additional data (e.g. taxonomic data, see below) this greatly enhances users capacity to interrogate the data generated by screening.
A MySQL client with a graphical user interface (e.g. SequelPro) can be used to connect to the screening database and select view the results of screening. For example, as shown below.
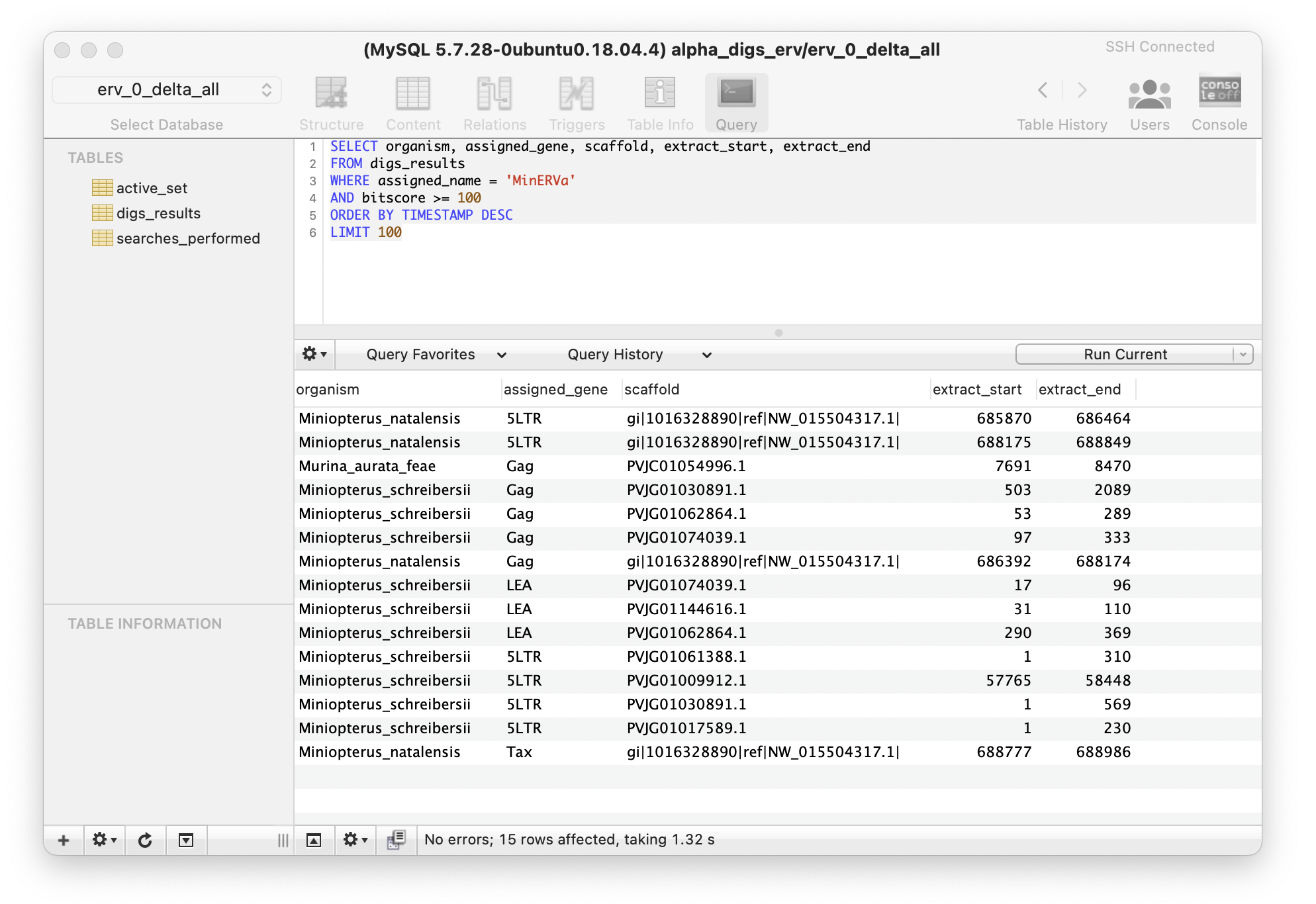
Visualising DIGS results: In the example shown above, an SQL statement is used to select sequences that matched to Miniopterus endogenous retrovirus (MinERVa) with a BLAST bitscore of at least 100. The 'ORDER BY' part of the SQL statement is used to sort the matching rows in order of the time they were entered into the results table (the TIMESTAMP field on each database table captures this information).
As well as selecting rows, it is often useful to count rows based using an SQL statement with a 'GROUP BY' clause, as shown in the example below.

Use of a 'GROUP BY' statement to count by category: In the example shown above, an SQL statement is used to select sequences that matched to Miniopterus endogenous retrovirus (MinERVa) with a BLAST bitscore of at least 100. The 'ORDER BY' part of the SQL statement is used to sort the matching rows in order of the time they were entered into the results table (the TIMESTAMP field on each database table captures this information).