-
Notifications
You must be signed in to change notification settings - Fork 2
Querying the GLUE Database

GLUE allows powerful queries to be run on the stored data, using a few simple concepts and command patterns. These can be used simply to extract data subsets, to ask simple research questions, or as part of a more complex analysis. To understand this section it is useful to be familiar with the core schema and schema extension sections. We will also use the example project for illustration so it will be useful to have this installed.
- GLUE "where clauses"
- Property paths to specify table columns
- Property paths in FreeMarker templates
Many GLUE commands have the same basic structure:
- Start from some particular set of candidate objects of the same type (i.e. in the same table)
- (Optionally) select a subset of candidate objects
- Perform some operation on the selected objects
GLUE "where" clauses are used in step 2 to select a subset of objects, using a logical filter. A command with this structure is the amino-acid frequency command. This command is executed in alignment mode, so a specific Alignment object is the data context.
Mode path: /
GLUE> project example
OK
Mode path: /project/example
GLUE> alignment AL_MASTER
OK
Mode path: /project/example/alignment/AL_MASTER
GLUE> amino-acid frequency -c -w "sequence.source.name = 'ncbi-hev-examples'" -r REF_MASTER_M73218 -f MT -l 56 66
+=======+===========+============+============+
| codon | aminoAcid | numMembers | pctMembers |
+=======+===========+============+============+
| 56 | V | 10 | 100.00 |
| 57 | F | 10 | 100.00 |
| 58 | R | 10 | 100.00 |
| 59 | P | 10 | 100.00 |
| 60 | E | 10 | 100.00 |
| 61 | V | 10 | 100.00 |
| 62 | L | 7 | 70.00 |
| 62 | F | 3 | 30.00 |
| 63 | W | 10 | 100.00 |
| 64 | S | 1 | 10.00 |
| 64 | N | 9 | 90.00 |
| 65 | H | 10 | 100.00 |
| 66 | P | 10 | 100.00 |
+=======+===========+============+============+
In this case the Alignment AL_MASTER is the data context, and the command operates as follows:
- The candidate object set consists of all AlignmentMembers which belong to Alignment AL_MASTER, or (since
-cwas used) one of its descendent Alignments in the alignment tree - Select a subset of these AlignmentMembers which pass the where clause filter specified by
-w "sequence.source.name = 'ncbi-hev-examples'" - Amongst the selected AlignmentMembers, compute the frequency of amino acid residues in the MT (Methyltransferase) genome region at ORF1 codon positions 56 to 66 inclusive (as specified by the
-r,-fand-loptions)
All the objects identified in step 1 will be tested in step 2 against the where clause; those which pass this filter will then contribute to the computation in step 3.
The where clause string sequence.source.name = 'ncbi-hev-examples' is supplied in double quotes in order to allow space and single quote characters to be used within the string.
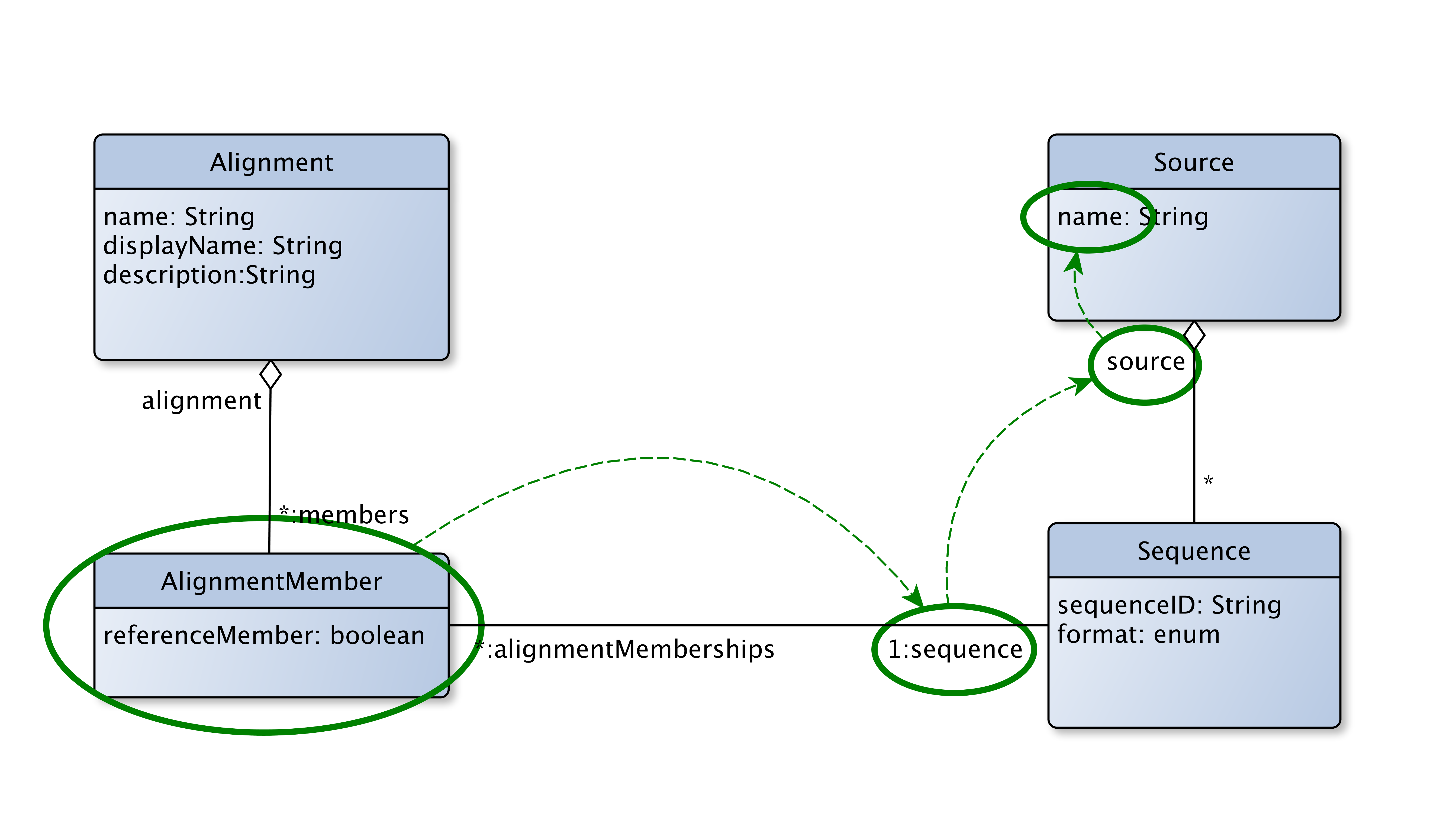
The first part of the where clause is sequence.source.name. This is a property path. Starting from the type of the candidate object, in this case AlignmentMember, the property path traverses named relational links specified by the data schema in order to arrive at a data value. In this case the all links are specified in the core schema; the relevant schema fragment is shown. However any or all of them could also be specified by a schema extension.
- Start from the AlignmentMember candidate object type
- Traverse the
sequencerelational link to the target Sequence object - Traverse the
sourcerelational link to the target Source object - Access the value for the field
nameon the Source object
The where clause expression uses the binary operator '=' to compare the Source name with the literal string 'ncbi-hev-examples', so that only AlignmentMembers based on Sequences belongs to Source ncbi-hev-examples are selected. It is also possible to use parentheses and logical operators such as 'and', 'not' and 'or' to join sub clauses together. Here are some where clause patterns you could use in the example project:
| Candidate object type | Where clause | Meaning | Note | Example command |
|---|---|---|---|---|
| Sequence | length >= 7200 and length < 7300 |
The field value for length is at least 7200 and less than 7300 |
Numeric comparison operators may be used for INTEGER fields such as length, DOUBLE or DATE fields |
list sequence |
| Sequence | length between 7200 and 7300 |
The field value for length is at least 7200 and at most 7300 |
The between ... and numeric comparison operator is for testing values within an inclusive range |
list sequence |
| m49_country | is_ldc = true or is_lldc = true |
Country is classified as least developed country or landlocked developing country | BOOLEAN fields such as is_ldc and is_lldc are tested for value true |
list custom-table-row |
| m49_country | m49_region.id = 'europe' and m49_sub_region.id != 'northern_europe' |
Country region is Europe but sub-region is not Northern Europe | The != comparison operator, which may be applied to field values of any type, is used for disequality, and the and logical operator conjoins two sub-clauses |
list custom-table-row |
| Sequence | gb_create_date > #gluedate(01-Apr-2009) |
First submitted to GenBank after 1st April 2009 | The #gluedate syntax specifies a concrete date, then numeric comparison operators may be used |
list sequence |
| Sequence | m49_country.id in ('CHN', 'JPN') |
Country of origin China or Japan | The in syntax allows comparison with any one of a fixed set of strings |
list sequence |
| Sequence | sequenceID like 'FJ%' |
The sequenceID field value starts with FJ |
The like operator my be used for String fields such as sequenceID. If so, the '%' character acts as a wildcard, matching any number of characters. |
list sequence |
| ReferenceSequence | alignmentsWhereRefSequence.name != null |
ReferenceSequence is the constraining reference sequence of at least one Alignment | The alignmentsWhereRefSequence link is specified in one of the core schema diagrams. When traversing from ReferenceSequence to Alignment the link allows multiple targets, so the where clause is effectingly requiring that there exists at least one link target |
list reference |
| Sequence | source.name = 'ncbi-hev-examples' and m49_country+.id = null |
Within Source ncbi-hev-examples but M.49 country is undefined | The keyword null can be used to test for an undefined value in the database. In this case we must also use the outer join operator + on link m49_country since by default sub-clauses which traverse to a non-existent link target will return false. The operator allows such sub-clauses to evaluate to null |
list sequence |
For those interested in implementation details, where clause strings are actually Apache Cayenne object path expressions.
The property path concept introduced above has other uses in GLUE. One example use is to configure the result columns of various "list ..." commands such as list custom-table-row, list sequence or list member. Such commands output a table where each row is an instance of a specific given object type. Each such command has a default set of columns, for example list member will give alignment.name, sequence.source.name and sequence.sequenceID. Note that these column headings are themselves property paths starting at the AlignmentMember object type. When invoking this command, you can supply your own property paths, overriding the columns in the result table, for example:
GLUE> alignment AL_MASTER
OK
Mode path: /project/example/alignment/AL_MASTER
GLUE> list member --recursive --whereClause "sequence.source.name = 'ncbi-hev-examples'" sequence.sequenceID alignment.name sequence.host_species
+=====================+================+=======================+
| sequence.sequenceID | alignment.name | sequence.host_species |
+=====================+================+=======================+
| AB481226 | AL_3e | - |
| AB591734 | AL_3a | Herpestes javanicus |
| AF444003 | AL_1b | - |
| FJ705359 | AL_3c | Sus scrofa |
| FJ763142 | AL_4a | Homo sapiens |
| FJ998015 | AL_3e | Sus scrofa |
| JF443717 | AL_1c | Homo sapiens |
| JQ013791 | AL_3ra | Oryctolagus cuniculus |
| JX855794 | AL_4b | Sus scrofa |
| KP294371 | AL_3i | Sus scrofa |
+=====================+================+=======================+
AlignmentMembers found: 10
Some GLUE modules use FreeMarker templates to assemble text strings. In some cases the model for the template may be a GLUE data object. Property paths may then be used to access the database from within the FreeMarker template.
A simple example is fastaAlignmentExporter. The ID string for each alignment row is formed using a FreeMarker template. The default behaviour is to use the following FreeMarker template to create an ID with three sections:
${alignment.name}.${sequence.source.name}.${sequence.sequenceID}
Each of the three sections is created by evaluating a property path from the AlignmentMember object that contributed the alignment row.
 GLUE by Robert J. Gifford Lab.
GLUE by Robert J. Gifford Lab.
For questions, issues, or feedback, please open an issue on the GitHub repository.
- Project Data Model
- Schema Extensions
- Modules
- Alignments
- Variations
- Scripting Layer
- Freemarker Templates
- Example GLUE Project
- Command Line Interpreter
- Build Your Own Project
- Querying the GLUE Database
- Working With Deep Sequencing Data
- Invoking GLUE as a Unix Command
- Known Issues and Fixes
- Overview
- Hepatitis Viruses
- Arboviruses
- Respiratory Viruses
- Animal Viruses
- Spillover Viruses
- Virus Diversity
- Retroviruses
- Paleovirology
- Transposons
- Host Genes