-
Notifications
You must be signed in to change notification settings - Fork 64
Parallelization
For impatient readers: How many threads should I use?
- Intro
- Adaptive parallelization
- Multithreading
- MPI and hybrid
- Thread pinning
- Core oversubscription
- Vector instructions
- How many threads?
- Coarse-grained parallelization
RAxML-NG supports three levels of parallelism: CPU instruction (vectorization), intra-node (multithreading) and inter-node (MPI). Starting with v.1.0.0, RAxML-NG supports both fine-grained parallelization across alignment sites (as in RAxML-PTHREADS and ExaML) as well as coarse-grained parallelization across tree searches (as in RAxML-MPI). Unlike with RAxML/ExaML, single RAxML-NG executable can handle all parallelism levels which can be configured in run-time (MPI support is optional and should be enabled at compile-time).
With fine-grained parallelization, the number of CPU cores that can be efficiently utilized is limited by the alignment "width" (=number of sites). For instance, using 20 cores on a single-gene protein alignment with 300 sites would be suboptimal, and using 100 cores would most probably result in a huge slowdown. In order to prevent the waste of CPU time and energy, RAxML-NG will warn you -- or, in extreme cases, even refuse to run -- if you try to assign too few alignment sites per core. Moreover, RAxML-NG v1.0.0 and later implements so-called adaptive parallelization, which liberates (unexperienced) users from dealing with technical details :)
By default, RAxML-NG will use a set of heuristics to determine the most efficient parallelization scheme given the specific dataset, analysis parameters and hardware configuration at hand:
-
For short alignment, RAxML-NG will automatically reduce the number of threads to prevent waste of resources.
-
If multiple tree inferences are requested (e.g., with
--searchor--allcommand), then RAxML-NG will consider running multiple searches ("workers") in parallel. It will automatically determine the "reasonable" number of workers based on total number of tree searches, alignment length, available memory, and number of CPU cores.
Importantly, user remains in full control and can override parallelization settings:
--threads 16 --workers 1 <- use *exactly* 16 threads, no coarse-grained parallelization
--threads 16 --workers auto <- use *exactly* 16 threads, heuristic will determine number of parallel tree searches
--threads auto{16} --workers auto{4} <- use *at most* 16 threads in total, split over *at most* 4 parallel tree searches
NOTE: The --threads option always specifies the overall number of threads per node, regardless of the number of workers! Hence, if you allocate full node(s) for your job, please use --threads auto or --threads auto{N}, where N is the number of physical CPU cores on your nodes!
At the moment, adaptive parallelization is based on a static heuristic, which is simple, very fast to compute, and yields reasonable estimates according to our tests. However, if you are willing to invest some effort into manual tuning for your specific dataset and system, you will almost certainly find an even better-performing parallelization setup. Alternatively, please check out our ParGenes pipelines which implements dynamic load-balancing and allows to infer thousands of gene trees with a single command.
By default, RAxML-NG will try to use as many threads as there are CPU cores available in your system (unless alignment is too short, see above). Most modern CPUs employ so-called hyperthreading technology, which makes each physical core appear as two logical cores to software. For instance, on my laptop with Intel i7-3520M processor, RAxML-NG will detect 4 (logical) cores and use 4 threads by default, even though this CPU has only 2 physical cores. Hyperthreading can be beneficial for some programs, but RAxML-NG shows best performance with one thread per physical core. Thus, I would recommend using the --threads option to set the number of threads manually, to be on the safe side (for short alignments, use --threads auto{MAX}).
If compiled with MPI support, RAxML-NG can leverage multiple compute nodes for a single analysis. Please check your cluster documentation for system-specific instructions for running MPI programs. In MPI-only mode, you should start 1 MPI process per physical CPU core (the number of threads will be set to 1 by default). However, in most cases, hybrid MPI/pthreads setup will be more efficient in terms of both runtime and memory consumption. Typically, you would start 1 MPI rank per compute node, and 1 thread per physical core (e.g. --threads 16 for nodes equipped with dual-slot octa-core CPUs).
Here is a sample job submission script for our home cluster using 4 nodes * 16 cores:
#!/bin/bash
#SBATCH -N 4
#SBATCH -B 2:8:1
#SBATCH --ntasks-per-node=1
#SBATCH --cpus-per-task=16
#SBATCH --hint=compute_bound
#SBATCH -t 08:00:00
raxml-ng-mpi --msa ali.fa --model GTR+G --prefix H1 --threads 16Once again, please consult your cluster documentation to find out how to properly configure a hybrid MPI/pthreads run. Please note that incorrect configuration can result in extreme slowdowns und hence waste of time and resources!
For attaining optimal performance, it is crucial to ensure that only one RAxML-NG thread is running on each physical CPU core. Usually, operating system can handle thread-to-core assignment automatically. However, some (misconfigured) MPI runtimes tend to pack all threads on a single CPU core, resulting in abysmal performance. To avoid this situation, each thread can be "pinned" to a particular CPU core.
In RAxML-NG, thread pinning is enabled by default only in hybrid MPI/pthreads mode when 1 MPI rank per node is used.
You can explicitly enable or disable thread pinning with --extra thread-pin and --extra thread-nopin, respectively.
A situation when multiple program threads have to share a single CPU core is sometimes called "oversubscription". Since RAxML-NG puts heavy load on CPUs pretty much all the time, core oversubscription is counter-productive and can lead to a major (>10x) slowdown. Because we repeatedly observed that oversubcription can go unnoticed by the users, wasting time and energy, we introduced an automatic oversubscription check starting with RAxML-NG version 1.0. If you see the error message
ERROR: CPU core oversubscription detected! RAxML-NG will terminate now to avoid wasting resources.
this might be due to one of the following reasons:
- You specified more threads than there are CPU cores in your system -> reduce number of threads
- You are using a shared server and some cores are occupied by other programs -> investigate / reduce number of threads
2b. A special case of the above: you submit a cluster job and request, e.g., 4 cores -> use
--threads auto{4} - You are (usually) using a hybrid MPI/pthreads parallelization, and MPI pinned RAxML-NG threads incorrectly -> check your cluster manual and submission script, try to use 1 MPI rank per node, try
--extra thread-pin/--extra thread-nopin - If and only if you excluded the possibilities 1-3 above, this could be a false alarm -> use
--force perf_threadsto disable the oversubscription check, make sure that RAxML-NG runtime is "reasonable", and please let us know
RAxML-NG will automatically detect the best set of vector instructions available on your CPU, and use the respective computational kernels to achieve optimal performance. On modern Intel CPUs, autodetection seems to work pretty well, so most probably you don't need to worry about this. However, you can force RAxML-NG to use a specific set of vector instructions with the --simd option, e.g.
raxml-ng --msa ali.fa --model GTR+G --simd sse
to use SSE3 kernels, or
raxml-ng --msa ali.fa --model GTR+G --simd none
to use non-vectorized (scalar) kernels. This option might be useful for debugging, since distinct vectorized kernel might yield slightly different likelihood values due to numerical reasons.
As always, simple questions are the toughest ones. You might as well ask: How fast should I drive?. In both cases, the answer would be: It depends. Depends on where you drive (dataset), your vehicle (system), and your priorities (time vs. money/energy). In RAxML-NG, we put some warning signs and radar speed guns, for your own safety. As in real life, you are free to ignore them (--force option), which can have two outcomes: (1) earlier arrival, or (2) lost time and money. Fortunately, unlike on the road, you can experiment with RAxML-NG safely, and I encourage you to do so.
A reasonable workflow for analyzing a large dataset would be as follows. First, run RAxML-NG in parser mode, e.g.
raxml-ng --parse --msa ali.fa --model partition.txt
This command will generate a binary MSA file (ali.fa.raxml.rba), which can be loaded by RAxML-NG much faster than the original FASTA alignment. Furthermore, it will print estimated memory requirements and the recommended number of threads for this dataset:
[00:00:00] Reading alignment from file: ali.fa
[00:00:00] Loaded alignment with 436 taxa and 1371 sites
Alignment comprises 1 partitions and 1001 patterns
NOTE: Binary MSA file created: ali.fa.raxml.rba
* Estimated memory requirements : 54 MB
* Recommended number of threads / MPI processes: 4
This recommended number of threads is computed using a simple heuristic and should yield a decent runtime/resource utilization trade-off in most cases. It is also a good starting point for your experimentation: you can now run tree searches with varying number of threads, e.g.
raxml-ng --search --msa ali.fa.raxml.rba --tree rand{1} --seed 1 --threads 2
raxml-ng --search --msa ali.fa.raxml.rba --tree rand{1} --seed 1 --threads 4
raxml-ng --search --msa ali.fa.raxml.rba --tree rand{1} --seed 1 --threads 8
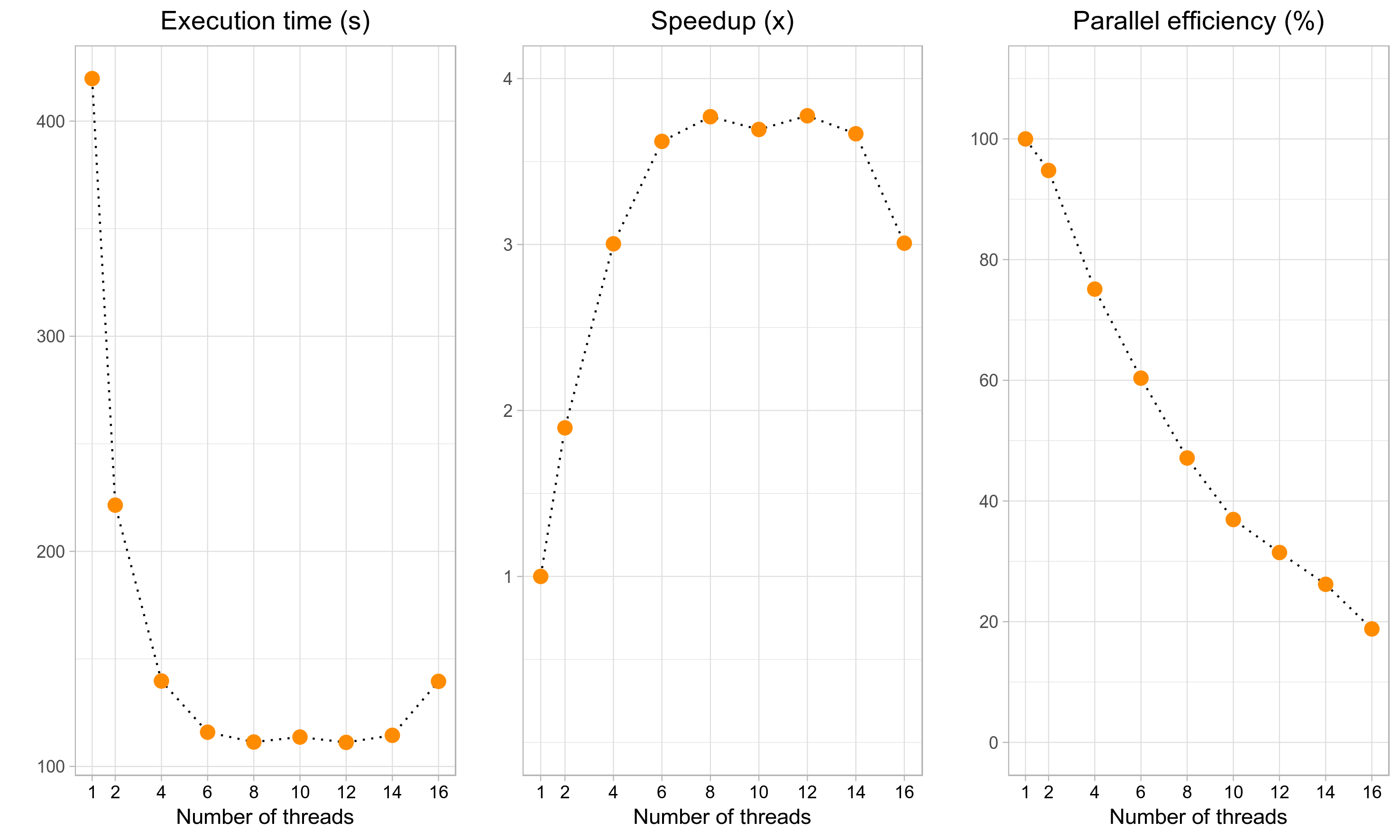
and so on. For a small dataset like in the example above, the execution time will decrease initially as we add more threads, but will then quickly level off (leftmost plot below). Although the maximum speedup of ~3.75x can be attained with 8-14 threads (middle plot), it corresponds to a rather poor parallel efficiency of 60%-30% (right plot). The recommended number of threads (4) yields a reasonable speedup (3x) without compromising the parallel efficiency too much (75%). Finally, if we use an excessively large number of cores (>=16 in this example), the execution time will start to increase again. Although the actual speedups will vary across datasets and systems, the general trend will stay the same. Therefore, it is up to the user to decide how much resources (=higher CPU time) can be sacrificed to obtain the results faster (=lower execution time).
In many practical scenarios, phylogenetic analysis comprises multiple tree inferences, e.g. ML search from distinct starting trees or using bootstrap replicate MSAs. In such cases, we can alleviate the parallelization bottleneck of the fine-grained approach by (additionally) parallelizing across independent tree searches. With RAxML-NG v1.0, using this coarse-grained parallelization is as simple as specifying the number of parallel tree searched via the --worker N option. Alternatively, you can rely on adaptive parallelization to determine the number of workers automatically.
Below we show results of a small benchmark that demonstrate the benefits of coarse-grained parallelization. We ran the --search command with 20 starting trees on a cluster node with 16 CPU cores, using varying number of workers, and measured following runtimes:
$ grep "Elapsed time:" *.raxml.log
T16_W1.raxml.log:Elapsed time: 2300.131 seconds <- fine-grained only
T16_W2.raxml.log:Elapsed time: 1255.979 seconds <- mixed, 2 workers
T16_W4.raxml.log:Elapsed time: 893.425 seconds <- mixed, 4 workers
T16_W8.raxml.log:Elapsed time: 902.180 seconds <- mixed, 8 workers
T16_W16.raxml.log:Elapsed time: 1044.757 seconds <- coarse-grained only, 16 workers (autodetect)
As we can see, using the optimal number of workers (4) yields speedup of ~2.6x compared to the fine-grained parallelization, whereas using the adaptive parallelization heuristic yields a somewhat lower - but still impressive - speedup of ~2.2x.