-
Notifications
You must be signed in to change notification settings - Fork 33
OortGA Fs
Although GO gene associations have traditionally been considered distinct from ontologies, there are advantages to putting gene association data on the same footing as the ontology
- integrated environment
- use of reasoners
- consistency checking
- querying
- deepening asserted classifications
- combining everything in an RDF triplestore
One particular use case for GO is to QC check the usage of relations in col16 (annotation extensions - a way of using anonymous class expressions in GO).
Another use case is using Oort to generate an ontology subset for a particular organism.
For GO you will need to load:
- a GAF (gzip format is fine)
- go/extensions/gorel.owl
- ro.owl
- go
You need to specify the GAF first, as this will be the "main" ontology from which to build the GO subset.
To do consistency checking, HermiT is currently the recommended reasoner.
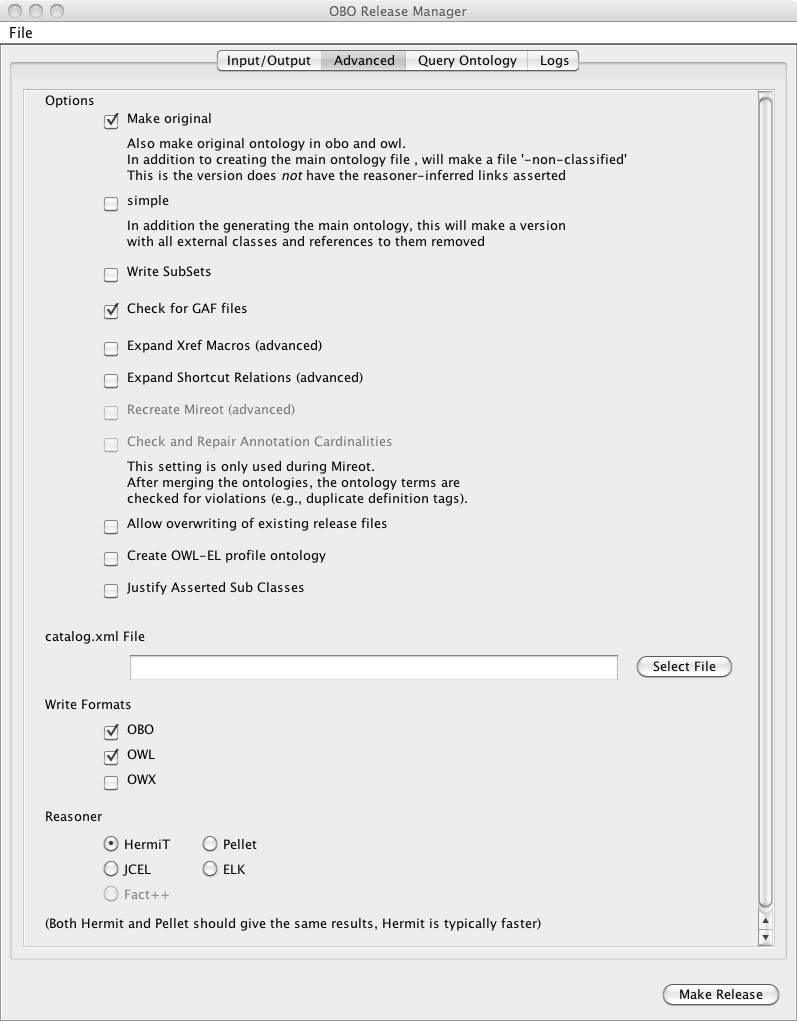
You should check the box that says "translate GAFs to OWL".
The Advanced Panel should look something like this:
[(for now it is best to have obo UNchecked)
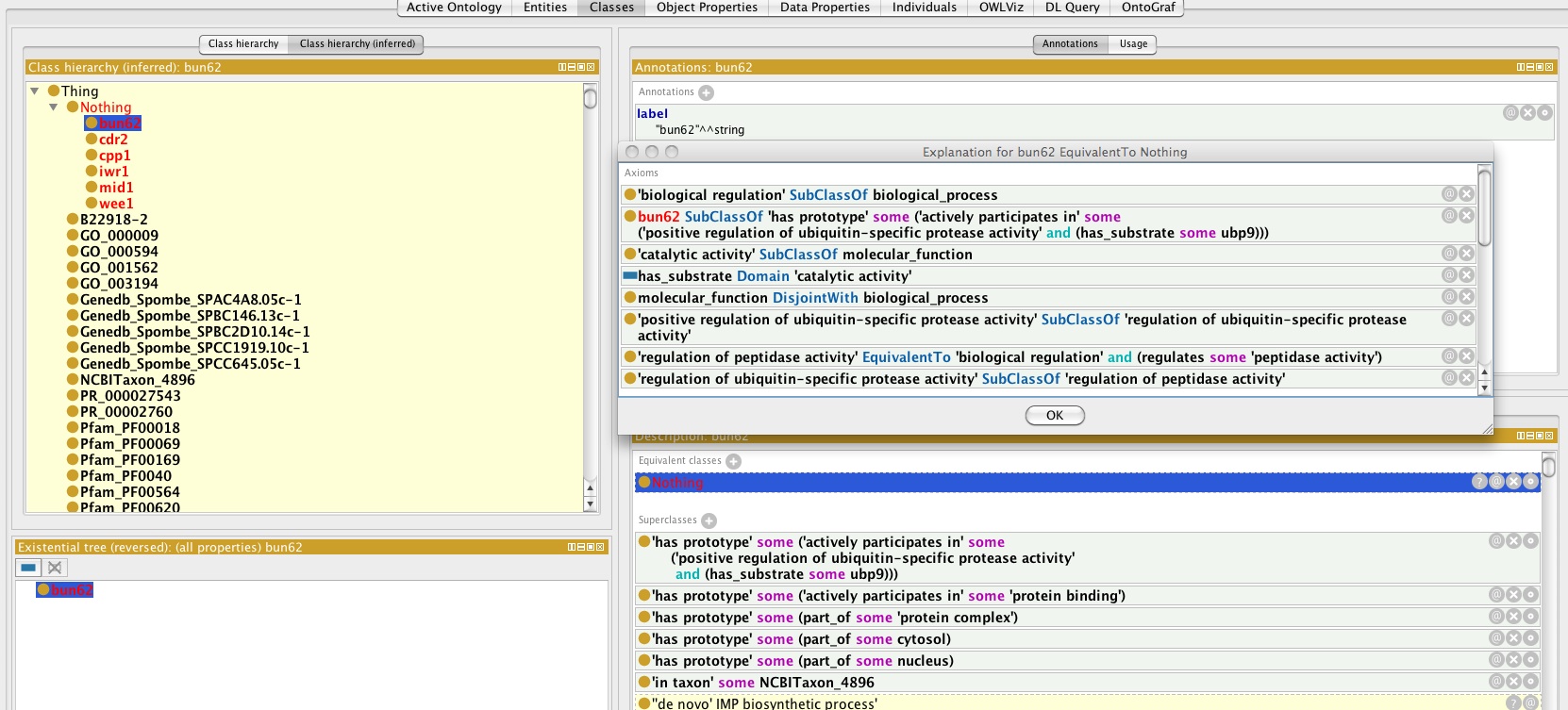
Oort will give you a somewhat mysterious message like "cpp1 is unsatisfiable". This means that given how you have described cpp1, there can be no instances of it (we treat genes as classes here).
In future Oort will have more detailed explanation messages. For now the best solution is to load the "merged.owl" file in Protege, invoke Hermit, find the unsatisfiable gene under "Nothing" and click on the "?" to get an explanation.
You should see something like:
- Annotation deepening
- GPAD support